(2: 南昌大学生命科学学院, 南昌 330031)
(2: School of Life Sciences, Nanchang University, Nanchang 330031, P. R. China)
蚌隶属软体动物门(Mollusca),双壳纲(Bivalvia),蚌目(Unionoida),包括3超科6科:Unionoidea (Unionidae + Margaritiferidae),Etherioidea (Mycetopodidae + Iridinidae + Etheriidae),Hyrioidea (Hyriidae)[1-2]. 作为淡水生物群落重要组成部分,蚌具有重要生态功能和经济价值[3-4]. 然而,受人类活动影响,该类群已成为最受威胁的动物群之一[5-6].
中国长江流域是东亚蚌类(蚌目:蚌超科)物种分布中心[1, 7]. Heude[8]曾依据贝壳形态特征描述中国蚌类140余种,奠定了我国蚌类分类基础. 此后,Simpson[9]以多种形态特征并将育儿囊类型作为属上阶元分类依据,记录中国蚌类14属,隶属蚌科(Unionidae) Unioninae和Hyriinae两个亚科. Haas[10]在Simpson研究基础上增加贝壳形态和钩介幼虫形态数据对其分类修订,记录中国蚌类20属,隶属蚌科Unioninae、Quadrulinae、Anodontinae和Lampsilinae四个亚科. 1950s开始,我国贝类学家做了大量区系调查和物种检索整理[4, 11-14],基于贝壳形态和解剖特征将我国蚌类分为蚌科珠蚌亚科(Unioninae)和无齿蚌亚科(Anodontinae),以及珍珠蚌科(Margaritiferidae)珍珠蚌属(Margaritiana)[4]. 到1990s,我国贝类学家重视软体部分解剖学和形态学特征,依据钩介幼虫形态和育儿囊类型,分类工作取得了一些进展和突破[15-18],但对中国蚌科属上阶元分类研究甚少,仍延续珠蚌亚科和无齿蚌亚科分类系统,而珍珠蚌科由于中国分布仅数种,其属上阶元分类鲜有探讨.
最近十几年,随着分子生物学技术的发展,基于贝壳形态学和解剖学分析与分子系统发育相结合的蚌目系统学研究得到快速发展,特别是蚌科系统发育框架已基本构建[19-24],但亚科阶元的系统发育关系仍有争议(图 1). 近几年的分子系统学研究通过增加分类单元促进了对蚌科高阶元系统发育的理解,但也存在亚科阶元重要节点处都出现低支持率现象,且得到的蚌科系统发育框架某些亚科并未形成单系群[21-24]. 世界上珍珠蚌科记录有效物种17种,广泛分布于世界各地,但其分布呈现明显不连续性和斑块性[2, 25]. 由于区域研究不平衡,该科分子系统研究最近几年才出现. Bolotov等[26]和Araujo等[27]基于多位点分子标记尝试解析该科系统发育关系,但并没有阐明高阶元系统分类. Lopes-Lima等[25]基于多分子标记和线粒体基因组学对该科提出2亚科4属分类系统.
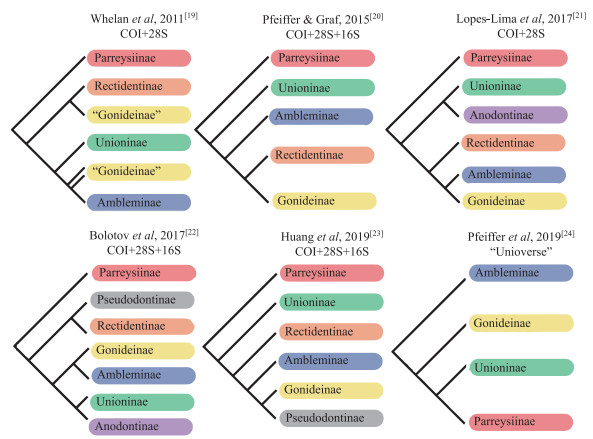
|
图 1 蚌科亚科阶元系统发育关系 Fig.1 Summary of recent phylogenetic hypotheses of subfamilies in the Unionidae |
中国蚌类分子系统学研究多聚焦于单属系统发育分析[28-29]或单种线粒体基因组学研究[30-32]. 少数对中国蚌类高阶元系统分类的研究[33-36],由于分类单元选择局限、系统发育分析方法单一、支序节点支持率低等原因导致很多属分类位置众说纷纭(表 1). 中国作为继北美另一蚌类物种分布中心[1-2, 7],补充其分类单元于当前蚌目系统发育框架,不仅能澄清中国蚌类物种分类地位,同时也能进一步发展完善世界蚌目系统发育关系.
| 表 1 中国蚌科属阶元分类历史 Tab. 1 Historical classification systems of Chinese genera in the Unionidae |
为此,本研究基于多位点分子标记(线粒体COI、16S rRNA、核基因28S rRNA)和线粒体基因组学,在蚌目系统发育框架下重构聚焦于世界蚌超科的系统发育关系,明晰中国蚌类物种分类地位和进化历史;同时检查和搜集蚌科14属物种贝壳形态学和解剖学特征,探讨蚌科属上阶元分类特征.
1 材料和方法 1.1 分类单元选择和数据集构建整理GenBank中所有蚌类序列,中国蚌超科当前(截止于2020年12月)可利用分子数据共计24属. 从GenBank中选择所有可利用的中国蚌超科分子序列,同时下载代表蚌超科各亚科分类单元的多位点分子数据和蚌超科所有F型线粒体基因组数据,构建聚焦于蚌超科的两个蚌目分子数据集:1)基于线粒体COI和16S以及核基因28S的多位点分子数据集;2)基于12蛋白质编码基因(除atp8)的线粒体基因组数据集. 最终,构建的多位点分子标记数据共计89种,其中中国蚌超科24属28种;构建的线粒体基因组数据集共计73种,其中中国蚌超科18属29种. 另选择三角蛤目(Trigoniida)海洋双壳类Neotrigonia margaritacea和Neotrigonia lamarckii为外类群(附表Ⅰ~Ⅱ).
1.2 序列比对和分区模型筛选对于多位点分子数据集,使用MEGA5.0[37]在无脊椎密码子翻译模式下将COI翻译成蛋白质序列,选择内置程序MUSCLE默认设置进行序列比对;使用MAFFT v7[38]选择Q-INS-i算法比对核糖体基因16S和28S. 利用Gblocks v0.91b[39]对模糊比对区域切割,设置参数核糖体基因Block最小长度为2 bp,允许缺失位点设置为With half;蛋白质编码基因Block最小长度为3 bp. 比对切割后的核苷酸序列利用SequenceMatrix[40]串联拼接,为后续系统发育分析构建多位点数据集(COI+16S+28S).
对于线粒体基因组数据集,利用PhyloSuite[41]从GenBank中批量下载所有选定的线粒体基因组数据,提取生成的核苷酸(NUC)数据包、氨基酸(AA)数据包以及基因排列(GO)文件. 使用MAFFT v7分别对NUC序列和AA序列比对,并利用Gblocks v0.91b删除比对中模糊对齐区域,移除所有缺失位点. 每个基因的NUC序列比对结果和AA序列比对结果分别保存,利用PhyloSuite串联生成后续系统发育分析的NUC数据集和AA数据集.
多位点数据集和两个线粒体基因组数据集基于编码基因和密码子分区处理:多位点数据执行5分区方案,NUC数据集执行36分区方案,AA数据集执行12分区方案. 在模型选择软件的统计标准中,BIC (bayesian information criterion)具有较高的准确性和精确性[42]. 因此利用PartitionFinder v1.1.1[43]和ModelFinder[44]基于最小BIC分别对多位点数据集、NUC数据集和AA数据集进行最适分区策略和模型筛选. 不同数据集分区策略和模型筛选见附表Ⅲ.
1.3 系统发育重建所有数据集,基于上述筛选的分区策略和模型构建BI (Bayesian inference)和ML (Maximum likelihood)系统发育分析. 在MrBayes v.3.2.6[45]中执行BI分析. 运行4条马尔可夫链,1百万代,每1000代抽样一次,burn-in 25 %,分裂频率平均标准差低于0.01时停止. 利用IQ-TREE[46]基于分区模型,运行1000次重复抽样检验,构建ML系统发育树. 利用在线iTOL(https://itol.embl.de/itol.cgi)实现系统发育树和基因排列文件的可视化和编辑.
利用MLGO (maximum likelihood for gene order analysis)[47]基于PhyloSuite生成的线粒体基因重排文件运行1000次重复抽样检验构建系统树,推测祖先基因排列. 利用CREx (common interval rearrangement explorer)[48]推测线粒体基因重排事件,包括基因反转(reversals)、换置(transpositions)、反向换置(reverse transpositions)、串联重复随机丢失(tandem duplication random loss, TDRL).
1.4 化石标定和分化时间估算基于多位点基因数据集利用BEAST v1.7.5[49]进行物种分化时间估算,构建物种进化树. 选择uncorrelated lognormal clock model作为分子钟模型,树的先验模型设定为birth-death speciation process. 利用PartitionFinder v1.1.1对数据分区替代模型筛选,结果见附表Ⅳ. MCMC代数设置为3亿代,取样频数为50000,设置不同分区替换模型为unlink. 利用Tracer 1.5估算收敛和有效取样大小,确保均大于200. 最后在TreeAnnotator v1.7.5软件中设置burnin 10 %,后验概率值限制为0.5,节点显示height median,生成最大谱系置信树.
参考先前研究,选择5个可靠的化石标定点分化时间:(1) 基于奥陶纪早期Noradonta Pojeta & Gilbert-Tomlinson, 1977[50],参考Huang等[23],对Palaeoheterodonta树的基部利用normal distribution prior限制为471~478 Ma (mean=475 Ma, stdev=2);(2) 最古老的化石Shifangella Liu & Luo, 1981可追溯至三叠纪晚期,参照Wu等[51],对珍珠蚌科起源时间标定230 Ma (exponential prior, lambda=30);(3) 蚌科最古老的化石发现在北美莫里逊组,参照Graf等[52],将蚌科最小分化时间设置为152 Ma (exponential prior, lambda=20);(4) 化石M. martinsoni,地质年代为渐新世和始新世间,参照Huang等[23],M. dahurica-M. margaritifera最近共同祖先最小设定为34 Ma (exponential prior, lambda=9.3);(5) 化石Lamprotula hungi,地质年代为渐新世和始新世间,参照Wu等[51],Lamprotula leaii和L. caveata最近共同祖先最小设定为34 Ma (exponential prior, lambda=9.3).
1.5 形态学和解剖学分析对保存于南昌大学生物博物馆的蚌科14属17种贝壳形态检查,包括贝壳形状、贝壳厚度、外套线和铰合部结构(附表Ⅴ).
综合前期研究和资料查阅,整理蚌科24属28种解剖学特征,包括钩介幼虫形态、育儿囊特征、繁殖特征、出入水孔特征等(附表Ⅵ).
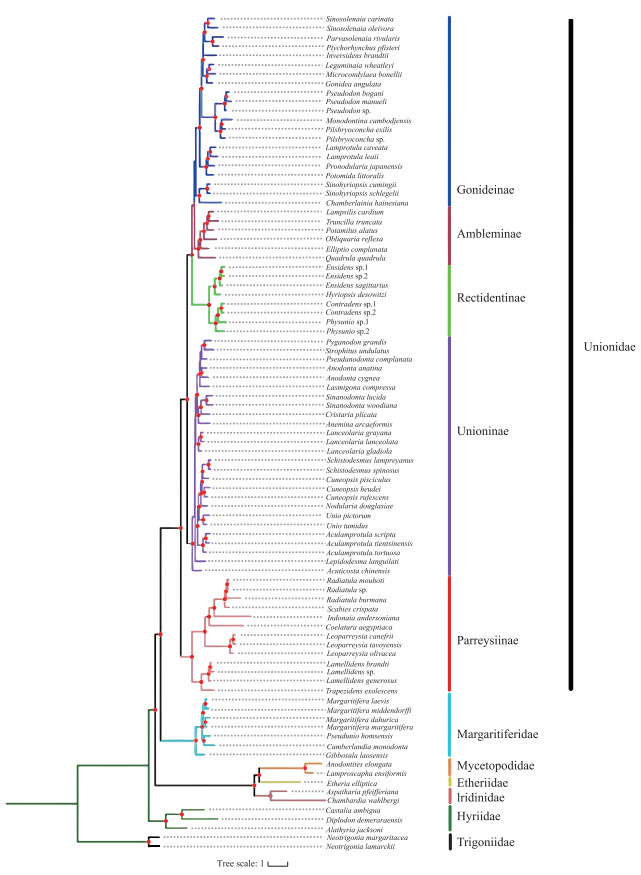
2 结果 2.1 基于多位点分子数据的蚌目系统发育分析基于多位点数据集构建的ML和BI树拓扑结构大体一致(图 2,附图Ⅰ),且绝大多数节点为强自展支持率(70 % ~100 %)和后验概率值(0.90~1.00).
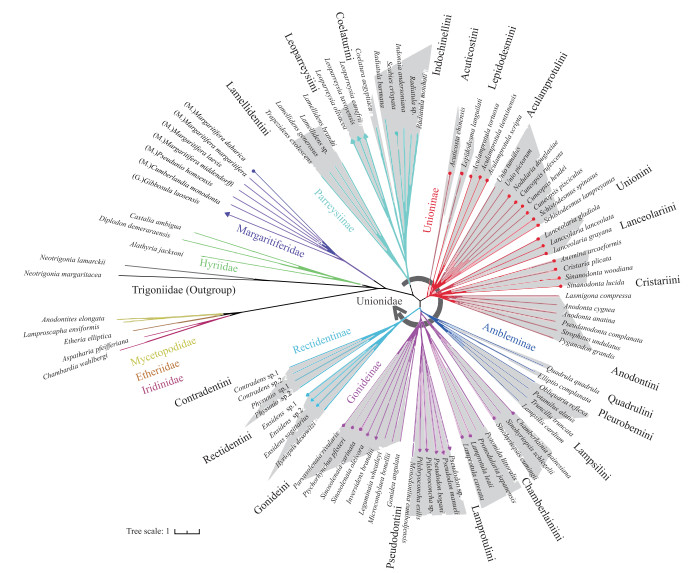
|
图 2 基于多位点数据集构建的蚌目系统发育树 (BI和ML树拓扑结构大体一致,这里仅展示BI树;图中不同颜色分枝代表蚌目不同科阶元以及蚌科亚科阶元,阴影部分代表蚌科族阶元;分枝末端圆点表示中国蚌种,三角表示中国蚌属;M表示珍珠蚌科珍珠蚌亚科,G表示珍珠蚌科弓背蚌亚科) Fig.2 Phylogenetic relationship of Unionoida based on three-gene dataset (Since the topologies of BI and ML trees are broadly consistent, only BI tree is shown here. Colored branches represent order levels of different families and subfamily levels of the Unionidae. Shaded sections represent tribe levels of the Unionidae. The circles at the end of the branch represent the Chinese unionid species, and the triangles represent the Chinese unionid genera. M. Margaritiferinae, G. Gibbosulinae) |
系统发育树显示蚌目各科系统发育关系如下:((Margaritiferidae + Unionidae) + (Iridinidae + (Etheriidae + Mycetopodidae))) + Hyriidae(图 2). 蚌科形成五个互为单系群亚科,亚科阶元系统发育关系如下:(((Ambleminae + Gonideinae) + Rectidentinae) + Unioninae) + Parreysiinae(图 2). 中国蚌超科24属隶属于珍珠蚌科(Margaritiferidae)和蚌科(Unionidae),其中珍珠蚌科中分属2个亚科(Gibbosulinae、Margaritiferinae);蚌科分属4亚科(Unioninae、Parreysiinae、Gonideinae、Rectidentinae) 13族(图 2).
2.2 蚌目线粒体系统发育基因组学基于线粒体NUC数据集和AA数据集共构建4棵拓扑结构完全一致的系统发育树(ML树2,BI树2),且绝大多数节点为100 % 自展支持率和1.00后验概率值(图 3).
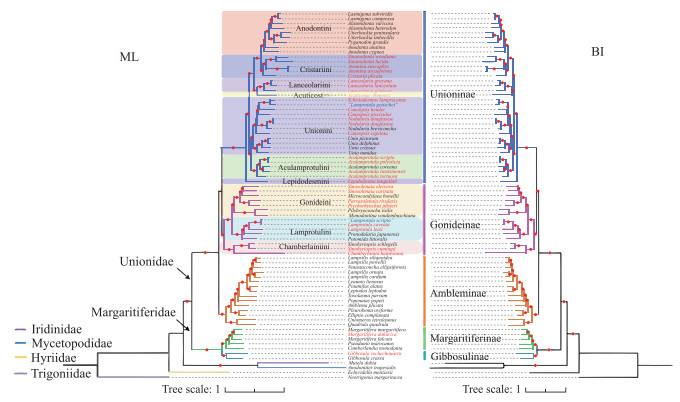
|
图 3 基于线粒体12蛋白质编码基因(除atp8)AA数据集和NUC数据集构建的ML和BI树 (进化枝红色圆点表示节点自展支持率(>70 %)和后验概率值(>0.9),红字字体表示中国蚌类物种. 注:由于AA数据集和NUC数据集构建的系统树拓扑结构一致,这里只展示AA数据集构建的系统树) Fig.3 Phylogenetic trees of freshwater mussels inferred from Bayesian Inference (BI) and Maximum Likelihood (ML) analyses (AA dataset of 12 mitochondrial protein-coding gene sequences (except atp8) were used reconstruction. Red circles on the branches represent bootstrap support (>70 %) and posterior probabilities (>0.9). Red font indicates Chinese species. Blue font indicates sequences in question. Since AA dataset and NUC data built completely congruent topologies, we only show the trees built by AA dataset here) |
系统发育树显示蚌目各科系统发育关系为:((Margaritiferidae + Unionidae) + (Iridinidae + Mycetopodidae)) + Hyriidae,蚌科互为单系群的3个亚科系统发育关系为:((Unioninae + Gonideinae) + Ambleminae).
中国蚌科(Unionidae)16属27种隶属2个亚科,即Unioninae和Gonideinae. Aculamprotula、Cuneopsis、Nodularia、Schistodesmus、Cristaria、Acuticosta、Lanceolaria、Sinanodonta、Anemina和Lepidodesma隶属于Unioninae;Lamprotula、Sinosolenaia、Sinohyriopsis、Chamberlainia、Parvasolenaia和Ptychorhynchus隶属于Gonideinae;中国珍珠蚌科(Margaritiferidae) 2属2种隶属2个亚科,即Margaritiferinae和Gibbosulinae(图 3).
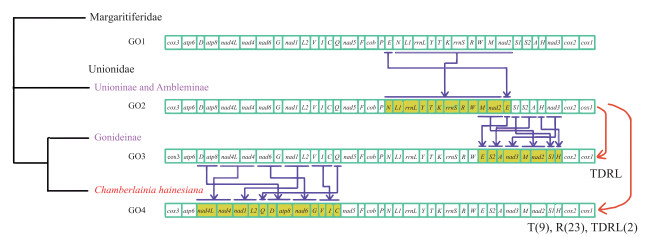
2.3 蚌超科线粒体基因组基因排列蚌超科(Unionoidea)线粒体基因组有4种基因排列方式(图 4). 珍珠蚌科(Margaritiferidae)独享一种基因排列方式GO1;蚌科(Unionidae) Unioninae和Ambleminae共享一种基因排列方式GO2;Gonideinae有2种基因排列方式GO3和GO4,其中室蚌(Chamberlainia hainesiana)独享GO4基因排列,其余Gonideinae物种共享GO3基因排列(图 4). 中国蚌超科物种线粒体基因组享有上述全部4种基因排列方式.
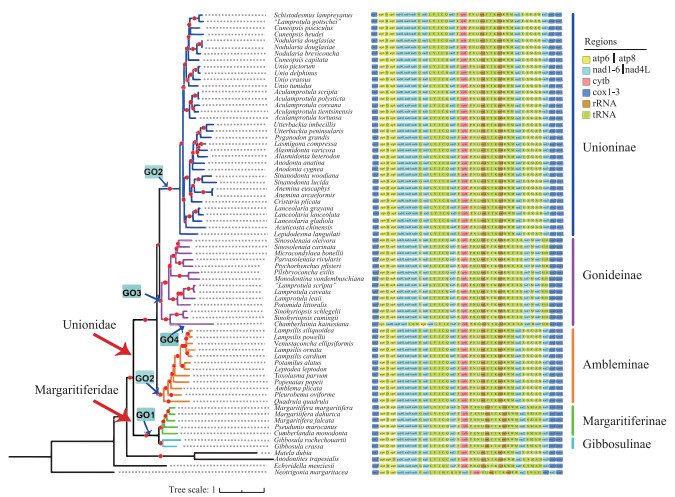
|
图 4 基于线粒体12蛋白质编码基因构建的系统发育树和相应的物种线粒体基因排列 Fig.4 Phylogenetic trees of freshwater mussels inferred from 12 protein-coding gene sequences and the corresponding mitochondrial gene order (GO) |
珍珠蚌科(Margaritiferidae)基因排列GO1与蚌科(Unionidae) Unioninae和Ambleminae基因排列GO2由于[trn N-nad2]基因块与trn E易位而发生重排. 基于线粒体基因重排构建的系统树显示:蚌科GO2位于GO3与GO4的基部,推测为蚌科祖先基因排列. GO3与祖先基因排列GO2基因重排主要发生在trn W和cox2间,由祖先基因排列经一次串联重复随机丢失事件进化而来. GO4与祖先基因排列GO2基因重排发生在atp6-nad5间,由祖先基因排列经多次基因反转、换置、以及串联重复随机丢失事件进化而来(图 5).
|
图 5 基于线粒体基因重排MLGO分析的系统树和CREx分析推定的基因重排事件 Fig.5 Phylogenetic trees inferred from MLGO analysis and putative gene rearrangement events based on gene order (T: transpositions; R: reverse; TDRL: tandem duplication random loss) |
利用BEAST基于多位点数据集构建的蚌目系统发育树,(Iridinidae + (Etheriidae + Mycetopodidae))位于蚌目基部进化枝(图 6). 蚌超科系统发育关系得到与BI和ML一致的拓扑结构,支持蚌科5亚科分类系统,即(((Ambleminae + Gonideinae) + Rectidentinae) + Unioninae) + Parreysiinae;珍珠蚌科2亚科分类系统,即(Gibbosulinae + Margaritiferinae)(图 6). 中国蚌超科隶属蚌科4亚科13族和珍珠蚌科2亚科(图 6).

|
图 6 基于多位点数据集化石标定的系统发育树 (节点条状表示95 % HPD,星号表示时间标定的化石,分枝末端圆点表示中国蚌种,三角表示中国蚌属,Ma:百万年前,Q.:第四纪,N.:第三纪) Fig.6 Fossil-calibrated phylogeny based on three-gene dataset (Node bars denote the 95 % highest posterior density (HPD). Fossils used for calibrations are marked by star signs. The circles at the end of the branch represent the Chinese unionid species, and the triangles represent the Chinese unionid genera. Ma: million years ago, Q.: Quaternary, N.: Neogene.) |
珍珠蚌科起源时间发生在晚石炭世(median=312.61 Ma, 95 % highest posterior density (HPD)=239.21~391.84),物种分化发生于晚白垩世(median=91.59 Ma, 95 % HPD=62.87~120.65). 蚌科物种分化时间发生在中三叠世(median=232.07 Ma, 95 % HPD=179.25~292.79),其中雕刻蚌亚科(Parreysiinae)最先起源,物种分化发生在早侏罗世(median=190.18 Ma, 95 % HPD=141.76~242.51),先于其他亚科;珠蚌亚科(Unioninae)起源时间在中侏罗世(median=181.23 Ma, 95 % HPD=140~230.02),物种分化发生于白垩纪(median= 114.36 Ma, 95 % HPD=85.4~152.77);矩形蚌亚科(Rectidentinae)起源于晚侏罗纪世(median=159.24 Ma, 95 % HPD=121.99~198.65),物种分化于晚白垩世(median=94.72 Ma,95 % HPD=66.47~125.69);小方蚌亚科(Ambleminae)和隆脊蚌亚科(Gonideinae)最近共同祖先分化于晚侏罗纪世(median=144.69 Ma,95 % HPD= 113.84~182.05). 中国蚌科物种最早分化时间可追溯到白垩纪(中国尖嵴蚌(Acuticosta chinensis), median=114.36 Ma, 95 % HPD=85.4~152.77).
3 讨论 3.1 蚌超科系统发育关系基于多位点分子标记和线粒体基因组学,本研究建立了蚌科5亚科分类系统,即(((Ambleminae + Gonideinae) + Rectidentinae) + Unioninae) + Parreysiinae,各亚科均得到节点支持率强的单系群分支. Huang等[23]通过对蚌科分类单元充分取样构建的蚌目系统发育树,由于选择分类单元可利用的分子标记不完整,导致蚌科亚科阶元分支节点支持率低,且Gonideinae为复系群. Pfeiffer等[24]利用锚定混合富集探针增加更多分子标记构建了蚌科4亚科分类系统,但分类单元取样有限,且亚科阶元分支支持率低. 本研究在当前具争议的亚科阶元中选择代表各亚科分类单元,综合取样单元完整的3分子标记重构蚌目系统发育关系,结果支持亚科阶元Pseudodontinae降级为族阶元Pseudodontini,同时支持亚科阶元Rectidentinae的有效性. 蚌科族阶元的系统分类近几年讨论较多,并且随着蚌科分类单元的增加,系统发育框架不断有新的族阶元被界定[21-23]. 本研究族阶元分类与Huang等[23]结果基本一致,但由于Cuneopsis、Nodularia、Schistodesmus、Unio形成单系群,故支持Pfeiffer等[24]将其归类于Unionini. 珍珠蚌科全球现存有效物种仅17种. 当前,该科2亚科4属分类系统在本研究中进一步得到佐证. 中国该科记录4种,即珠母珍珠蚌Margaritifera dahurica (Middendorff, 1850)、粗糙弓背蚌Gibbosula confragosa(Frierson, 1928)、佛耳弓背蚌Gibbosula mansuyi (Dautzenberg & Fischer,1908)和猪耳弓背蚌Gibbosula rochechouartii(Heude, 1875),隶属珍珠蚌亚科(Margaritiferinae)和弓背蚌亚科(Gibbosulinae).
在BEAST分子钟中,珍珠蚌科起源和分化时间出现221 Ma的长分枝现象,这可能是祖先亲缘关系近的分类群在古环境压力下灭绝所致,最古老的化石记录在侏罗纪中期和晚期也支持这一假设[26]. 由于这种灭绝,该科物种分化时间可能会更早. 然而,现生珍珠蚌分化却可追溯到白垩纪晚期91 Ma,这与先前研究取得一致(mean age=88.33 Ma)[53]. 本研究蚌科分化时间与Huang等[23]取得基本一致. 长江流域独特且多样的生态系统孕育了我国物种多样性丰富、特有程度高、远古物种多的蚌类,我国蚌科物种分化时间集中发生于晚白垩纪和古近纪(36~100 Ma). 中国特有物种中国尖嵴蚌(Acuticosta chinensis)可能是蚌超科最古老的现生物种,其物种起源可追溯至白垩纪(mean age=114.36 Ma).
此外,对GenBank所有蚌超科线粒体全基因组搜集重构系统发育关系时发现,Lamprotula scripta (NC_030258)与Lamprotula物种聚为一支,Lamprotula gottschei (NC_023806)与Schistodesmus lampreyanus形成姐妹群,本研究对这2条序列提出质疑. Lamprotula scripta已有多种分子数据证实该物种隶属Aculamprotula,对其重新采样并线粒体基因组测序(NC_045529),也证实该物种分类位置为Aculamprotula[54-55]. 另Lamprotula gottschei与Schistodesmus lampreyanus形成姐妹群,推测可能是样本鉴定有误或序列测定过程DNA污染所致.
3.2 中国蚌超科属阶元分类系统本研究构建了目前最为全面的中国蚌超科系统发育关系,明确了中国蚌超科24属系统分类地位(表 2). 结合历史相关资料,整理得中国蚌超科共计26属. 由于假柏夫蚌属(Pseudobaphia)和菱形蚌属(Rhombuniopsis)缺乏分子数据,暂未对其做高阶元分类(表 2中*表示).
| 表 2 中国蚌超科属阶元分类系统 Tab. 2 Classification system of Chinese Unionoidea |
本研究对中国蚌科属级阶元分歧较大的属,如中国蛏蚌属(Sinosolenaia)、中国帆蚌属(Sinohyriopsis)、结节蚌属(Nodularia)、中国无齿蚌属(Sinanodonta)和船蚌属(Anemina)系统评述,阐明争议属分类历史.
Subfamily Gonideinae Ortmann, 1916
Genus Sinosolenaia Bolotov, Kondakov, Konopleva & Vikhrev, 2021
Type species: Sinosolenaia recognita (Heude, 1877)
Species: Sinosolenaia oleivora (Heude, 1877) 图 7A

|
图 7 中国蚌类贝壳形态 (A:中国橄榄蛏蚌,B:中国三角帆蚌,C:短褶矛蚌,D:扭矛蚌,E:蚶形船蚌,F:中国背角无齿蚌,G:圆顶结节蚌) Fig.7 Shell morphology in Chinese unionids (A: Sinosolenaia oleivora, B: Sinohyriopsis cumingii, C: Lanceolaria grayanus, D: Lanceolaria lanceolata, E: Anemina arcaeformis, F: Sinanodonta woodiana, G: Nodularia douglasiae) |
Comments: 该属为中国特有属. 属内物种基于贝壳形态长期归类于泰国分布的以Solenaia emarginata (Lea, 1860)为模式种的蛏蚌属(Solenaia). 随着蚌科分子序列增加,最近系统发育分析发现中国蛏蚌属与分布于泰国的蛏蚌属隶属不同亚科,中国蛏蚌属隶属Gonideinae,而泰国蛏蚌属隶属Rectidentinae[22, 24]. Bolotov等[56]提议中国分布的该类群应另立一属,即Sinosolenaia.
Description: 贝壳窄长似蛏形,壳前部细圆,狭长,后部逐渐向延长扩大,呈斜截状. 壳顶低,不突出背缘之上,常被腐蚀[4]. 外套线明显,铰合部甚弱,无主齿,仅有弱的侧齿痕迹(附表Ⅴ). 无真正鳃水管和隔膜;育儿囊有内外4片鳃瓣构成,无胶质索形成,属内物种钩介幼虫形态存在差异(附表Ⅵ).
Distribution: 该属物种为中国特有种,分布于长江中下游地区[4],其中中国龙骨蛏蚌(S. carinata)现存仅分布于江西鄱阳湖流域[57].
Genus Sinohyriopsis Starobogatov, 1970
Type species: Sinohyriopsis cumingii (Lea, 1852) 图 7B
Comments: 属内记录2种,即Sinohyriopsis cumingii和Sinohyriopsis schlegelii. S. cumingii为中国本地种,S. schlegelii为中国引进种[58]. 基于贝壳形态以往该属物种归类于Hyriopsis. 直到现在国内一些分类学和生态学研究仍沿用Hyriopsis,特别是一些区域物种多样性调查[59-61]. 最近很多分子系统研究,包括本研究已证实,Hyriopsis隶属Rectidentinae,主要集中在东南亚地区,包括东印度、缅甸、泰国、老挝、越南、柬埔寨、马来西亚半岛、印度尼西亚,而Sinohyriopsis则隶属Gonideinae,主要分布在中国和日本地区[21-22, 24].
Description: 贝壳呈三角形,大型,侧扁,壳质较厚;前端低,后端高;后背缘向上突起形成三角形的翼[4];外套线明显,铰合部发达;壳顶不突出背缘之上(附表Ⅴ). 育儿囊为外鳃型,钩介幼虫无钩,无胶质索形成(附表Ⅵ).
Distribution: 该属物种广泛分布在中国长江和黄河流域,以及日本和越南[21].
Subfamily Unioninae Rafinesque, 1820
Genus Lanceolaria Conrad, 1853
Type species: Lanceolaria grayii (Griffith & Pidgeon, 1833)
Species: Lanceolaria grayanus (Lea, 1834) 图 7C
Lanceolaria lanceolata (Arconaia lanceolata) (Lea, 1856) 图 7D
Comments: 该属长期基于形态学研究归入Unioninae. 后来,Lopes-Lima等[25]和Wu等[36]基于分子数据支持该属归类于Anodontinae. 最近,随着Anodontinae并到Unioninae,该物种重新归类于Unioninae. Arconaia为中国特有属,仅包含一种,即Arconaia lanceolate (Lea, 1856). 由于Arconaia与Lanceolaria物种具有更近的近缘关系,并形成单系群,Huang等[23]基于分子数据将Arconaia合并到Lanceolaria,Arconaia lanceolata重组为Lanceolaria lanceolate comb. res.
Description: 贝壳为细长型;壳质较厚,壳顶低[4];外套线明显,铰合齿发达(附表Ⅴ). 有真正鳃水管,瓣间隔膜完整;育儿囊为外鳃型,钩介幼虫呈三角形,有钩,属内钩介幼虫在育儿囊中发育方式不同(附表Ⅵ).
Distribution: 该属物种主要分布于远东亚,包括俄罗斯、日本、韩国、朝鲜、中国长江流域、越南[21].
Genus Anemina Haas, 1969
Type species: Anemina arcaeformis (Heude, 1877) 图 7E
Comments: 见Genus Sinanodonta.
Description: 壳质薄,易碎;外形呈椭圆形,壳顶膨胀,突出于背缘之上;壳面呈淡黄绿色或黄褐色,有光泽,具有细弱同心圆的生长轮脉[4];铰合部弱,无齿;外套线不明显(附表Ⅴ). 有真正鳃水管和隔膜,入水管有2~3行排列乳突,出水管光滑无乳突;育儿囊为外鳃型,只有外鳃瓣构成;钩介幼虫呈半圆形,有壳钩,无胶质索形成(附表Ⅵ).
Distribution: 同Sinanodonta分布.
Genus Sinanodonta Modell, 1944
Type species: Sinanodonta woodiana (Lea, 1834) 图 7F
Comments: Anemina以Anemina arcaeformis作为模式种而命名,Sinanodonta则以Sinanodonta woodiana作为模式种而命名. 由于贝壳形态相似,以往Sinanodonta和Anemina物种均归类于Anodonta. 直到现在国内一些分类学和生态学研究仍沿用Anodonta,特别是在一些区域多样性调查中[59-61]. 如本研究所示,Anodonta隶属Anodontini;Sinanodonta和Anemina为两个独立单系群,均隶属Cristariini[23]. 这两族有明显地域分布差异,Anodonta主要分布于北美和西古北界地区,而中国则只有Sinanodonta和Anemina分布[21].
Description: 壳质薄,易碎;外形呈椭圆形,壳顶不膨胀或稍膨胀,不突出于背缘之上;壳面呈淡黄绿色或黄褐色,有光泽,具有细弱同心圆的生长轮脉[4];铰合部弱,无齿;外套线不明显(附表Ⅴ). 有真正鳃水管和隔膜,入水管有2~3行排列的乳突,出水管光滑无乳突;育儿囊为外鳃型,钩介幼虫呈长三角形,无胶质索形成(附表Ⅵ).
Distribution: 广泛分布于印度支那半岛到中国、朝鲜、韩国、日本以及俄罗斯、北美等地[21].
Genus Nodularia Conrad, 1853
Type species: Nodularia douglasiae (Griffith & Pidgeon, 1833) 图 7G
Comments: 该属以Nodularia douglasiae为模式种而建立. 以往我国该属物种归类于Unio,现在一些区域多样性调查仍沿用Unio douglasiae[59-61]. 当前已有很多分子数据证实Nodularia和Unio为两个独立单系群,同时两属具有明显地域分布差异,Unio主要分布于非洲热带、东亚、北美和欧亚大陆北部,Nodularia仅分布于东亚,而目前中国仅发现Nodularia分布[21, 62].
Description: 贝壳卵形或长椭圆形[4];壳质较厚,外套线明显,铰合齿发达(附表Ⅴ). 有鳃水管,瓣间隔膜不完整;入水管有2~3行排列的乳突,出水管有乳突;育儿囊由外鳃瓣构成,钩介幼虫呈三角形,有钩,有胶质索(附表Ⅵ).
Distribution: 广泛分布在中国(包括台湾)、韩国、马甲丹、俄罗斯日本和库页岛[21].
3.4 形态学和解剖学特征在蚌科属上阶元分类中的意义以往蚌科分类特征主要包括以下方面:贝壳形态、软体部分形态、钩介幼虫形态以及某些繁殖习性. 早期Simpson[9]基于贝壳形态、钩介幼虫形态和育儿囊类型将全球蚌科分为Unioninae和Hyriinae两个亚科. 后来Ortmann[63]指出由于贝壳形态趋同性大,以其作为科及亚科阶元分类特征不适当,并综合其他解剖特征将北美蚌科分为Unioninae、Anodontinae、Lampsilinae三个亚科. 此后Modell[64]和Haas[10]基于形态学和解剖学特征对蚌科提出不同分类系统,但很大程度仍依赖贝壳形态. Heard等[65]则强调解剖学和繁殖特征差异,首次将繁殖特征作为科阶元分类依据.
贝壳形态特征易受生物和非生物双重影响,如年龄、大小、性别以及生境、水文、理化因素等[66-68]. 仅依靠或侧重于依赖贝壳形态对蚌科属上阶元分类不明智. 本研究也发现壳质、形状、铰合齿等贝壳特征并不能作为蚌科属上阶元分类依据(附表Ⅱ). 以往钩介幼虫形态和育儿囊特征是蚌科属及属上阶元重要分类依据[15-16, 64-65]. 然而,钩介幼虫形态不仅在属间存有差异,在属内也有差异. Sinosolenaia carinata和Sinosolenaia oleivora软体部分及钩介幼虫形态有明显区别(附表Ⅲ):S. oleivora进水管乳突为多分支状,出水管光滑,而S. carinata进水管和出水管均有乳突,但其乳突并没有像S. oleivora呈现尖锐状;S. oleivora钩介幼虫无钩中等大小,而S. carinata钩介幼虫则有钩且小,且繁殖期也大不相同. 这种属内幼虫形态差异在Pseudodon也有报道[20]. 同样,育儿囊类型不仅在属上阶元存在差异,属内种间也有差异,如Hyriopsis bialata和Hyriopsis desowitzi(附表Ⅲ).
因此,贝壳形态和解剖特征难以作为诊断各亚科的分类依据. Heard等[65]基于解剖学和繁殖特征数据构建北美蚌类系统发育,发现蚌类高阶元分子系统进化关系与特征演化史并不一致. 本研究对中国蚌科争议属形态描述仅是基于所搜集的贝壳形态和解剖特征,并不能作为该属分类诊断依据,不排除属内其他物种出现描述之外的其他特征. Zanatta等[69]基于分子数据探讨了Lampsilini族主动寄生策略出现与消失的演化过程. 或许今后研究可以尝试在系统发育框架上综合更多形态解剖学数据,包括生态特征和行为特征来探讨物种特征演化史,了解物种演化特征与生活史策略的适应性关系.
4 结论基于多位点分子标记(COI、16S、28S)和线粒体基因组学重构聚焦于蚌超科(蚌科和珍珠蚌科)的蚌目系统发育关系,结果支持蚌科5亚科分类系统,即(((Ambleminae + Gonideinae) + Rectidentinae) + Unioninae) + Parreysiinae;珍珠蚌科2亚科分类系统,即(Gibbosulinae + Margaritiferinae). 中国蚌类24属隶属蚌科4亚科(Gonideinae、Rectidentinae、Unioninae、Parreysiinae) 13族和珍珠蚌科两亚科(Gibbosulinae、Margaritiferinae). 蚌科物种分化时间为中三叠世,其中雕刻蚌亚科(Parreysiinae)起源和物种分化时间早于其他亚科;矩形蚌亚科(Rectidentinae)物种分化时间最晚,为晚白垩世;小方蚌亚科(Ambleminae)和隆脊蚌亚科(Gonideinae)最近共同祖先分化于晚侏罗纪世. 珍珠蚌科起源于晚石炭世,物种分化于晚白垩世. 中国尖嵴蚌(Acuticosta chinensis)可能是中国现生蚌科中起源最早的物种,可追溯到白垩纪(median=114.36 Ma).当前蚌超科线粒体基因组有4种基因排列方式,珍珠蚌科独享一种;蚌科中Unioninae和Ambleminae共享一种;Gonideinae有2种基因排列方式,其中室蚌(Chamberlainia hainesiana)独享一种. 以往作为高阶元分类的贝壳形态、钩介幼虫形态以及育儿囊类型,并不能作为蚌科属及以上阶元分类依据.
5 附录附表Ⅰ~Ⅵ和附图Ⅰ见电子版(DOI: 10.18307/2021.0615).
| 附表Ⅰ 本研究用于构建多位点联合数据集的序列号 Appendix Ⅰ List of sequences used in this study |
| 附表Ⅱ 用于系统发育分析的mtDNA数据集和对应登录号* Appendix Ⅱ mtDNA sequence used for molecular analyses and corresponding GenBank numbers |
| 附表Ⅲ 基于Moder Finder和Partition Finder对多位点联合数据集,线粒体AA数据集和NUC筛选的分区方案和最适模型 Appendix Ⅲ Partitioning srtategies from Moder Finder and Partition Finder for three locus dataset, mt AA dataset and mt NUC dataset |
| 附表Ⅳ 利用Partitionfinder筛选的最适分区和模型 Appendix Ⅳ Best partitioning scheme and model using partition finder |
| 附表Ⅴ 蚌科物种贝壳形态特征* Appendix Ⅴ Shell morphological characters in Unionidae |
参考文献
[1] Lopes-Lima M, Froufe E, Ghamizi M et al. Phylogeny of the most species-rich freshwater bivalve family (Bivalvia: Unionida: Unionidae): Defining modern subfamilies and tribes. Molecular Phylogenetics and Evolution, 2017, 106: 174-191.
| 附表Ⅵ 蚌科物种解剖形态特征* Appendix Ⅵ Anatomical morphological features in Unionidae |
参考文献
[1] Wu XP. Freshwater shellfish in the middle and lower reaches of the Yangtze River[Dissertation]. Wuhan: Institute of Hydrobiology, Chinese Academy of Sciences, 1998.[吴小平. 长江中下游淡水贝类的研究[学位论文]. 武汉:中国科学院水生生物研究所, 1998.]
[2] Cao Y, Liu X, Wu R et al. Conservation of the endangered freshwater mussel Solenaia carinata (Bivalvia, Unionidae) in China. Nature Conservation, 2018, 26 : 33-53.
[3] Lopes-Lima M, Froufe E, Ghamizi M et al. Phylogeny of the most species-rich freshwater bivalve family (Bivalvia: Unionida: Unionidae): Defining modern subfamilies and tribes. Molecular Phylogenetics and Evolution, 2017, 106 : 174-191.
[4] Wang Y, Zhang G, Wei K et al. Reproductive traits of the threatened freshwater mussel Solenaia oleivora (Bivalvia: Unionidae) from the middle Yangtze River. Journal of Molluscan Studies, 2015, 81(4): 522-526.
[5] Wu R, Chen T, Zanatta DT et al. Reproductive traits of nine freshwater mussel species (Mollusca: Unionidae) from Poyang Lake, China. Journal of Molluscan Studies, 2018, 84(3): 324-332.
|
附图Ⅰ 基于多位点分子标记构建的蚌目ML树 (进化枝红色圆点表示节点自展支持率(>70 %),图中不同颜色的分枝代表蚌目不同科阶元以及蚌科亚科阶元) AttachedFigⅠ Phylogenetic trees of freshwater mussels inferred from Bayesian Inference (BI) analyses (Red circles on the branches represent bootstrap support (>70 %) Colored branches represent order levels of different families and subfamily levels of the Unionidae) |
| [1] |
Bogan AE. Global diversity of freshwater mussels (Mollusca, Bivalvia) in freshwater. Hydrobiologia, 2008, 595(1): 139-147. DOI:10.1007/s10750-007-9011-7 |
| [2] |
Graf DL, Cummings KS. Review of the systematics and global diversity of freshwater mussel species (Bivalvia: Unionoida). Journal of Molluscan Studies, 2007, 73(4): 291-314. DOI:10.1093/mollus/eym029 |
| [3] |
Vaughn CC, Spooner DE. Unionid mussels influence macroinvertebrate assemblage structure in streams. Journal of the North American Benthological Society, 2006, 25(3): 691-700. DOI:10.1899/0887-3593(2006)25[691:umimas]2.0.co;2 |
| [4] |
Liu YY. Economic animal of China: Freshwater molluscs. Beijing: Science Press, 1979. [刘月英. 中国经济动物志: 淡水软体动物. 北京: 科学出版社, 1979.]
|
| [5] |
Lydeard C, Cowie RH, Ponder WF et al. The global decline of nonmarine mollusks. BioScience, 2004, 54(4): 321-330. DOI:10.1641/0006-3568(2004)054[0321:TGDONM]2.0.CO;2 |
| [6] |
Bogan AE. Freshwater bivalve extinctions (Mollusca: Unionoida) search for causes. Integrative and Comparative Biology, 1993, 33(6): 599-609. DOI:10.1093/icb/33.6.599 |
| [7] |
Zieritz A, Bogan AE, Froufe E et al. Diversity, biogeography and conservation of freshwater mussels (Bivalvia: Unionida) in East and Southeast Asia. Hydrobiologia, 2018, 810(1): 29-44. DOI:10.1007/s10750-017-3104-8 |
| [8] |
Heude P. Conchyliologie fluviatile de la Province de Nanking et de la Chine centrale. Paris: Savy, 1875. DOI:10.5962/bhl.title.14450 |
| [9] |
Simpson CT. Synopsis of the Naiades: or pearly fresh-water mussels. US Government Printing Office, 1900. |
| [10] |
Haas F ed. Superfamilia unionacea. de Gruyter, 1969.
|
| [11] |
Zhang X, Li SC. Bivalves from Poyang Lake and its surrounding waters include a new species. Acta Zoologica Sinica, 1965, 17(3): 309-317. [张玺, 李世成. 鄱阳湖及其周围水域的双壳类包括一新种. 动物学报, 1965, 17(3): 309-317.] |
| [12] |
Lin ZT. Unionidae (Mollusca) of Poyang Lake, Jiangxi province, China. Acta Zoologica Sinica, 1962, 14(2): 249-260. [林振涛. 鄱阳湖的蚌类. 动物学报, 1962, 14(2): 249-260.] |
| [13] |
Liu YY, Zhang WZ, Wang YX. Bivalves (Mollusca) of the Taihu and its surrounding waters, Jiangsu province, China. Acta Zoologica Sinica, 1980(4): 365-369. [刘月英, 张文珍, 王跃先. 太湖及其周围水域的双壳类. 动物学报, 1980(4): 365-369.] |
| [14] |
Liu YY, Zhang WZ, Wang YX et al. New records of Chinese freshwater clams. Acta Zootaxonomica Sinica, 1979, 4(2): 188. [刘月英, 张文珍, 王耀先等. 我国蚌类新纪录. 动物分类学报, 1979, 4(2): 188.] |
| [15] |
Wu XP, Liang YL, Wang HZ. A comparative study on glochidial morphology of Unionidae (Bivalvia) I. Unio douglasiae, Cuneopsis pisciulus, Acuticosta chinensis and Acuticosta ovata. Acta Hydrobiologica Sinica, 1999, 23(2): 141-145. [吴小平, 梁彦龄, 王洪铸等. 蚌科钩介幼虫比较形态学研究——Ⅰ. 四个种幼虫的形态. 水生生物学报, 1999, 23(2): 141-145. DOI:10.3321/j.issn:1000-3207.1999.02.007] |
| [16] |
Wu XP, Liang YL, Wang HZ et al. A comparative study on glochidial morphology of Unionidae (Bivalvia). Acta Hydrobiologica Sinicaz, 2000, 24(3): 252-256. [吴小平, 梁彦龄, 王洪铸等. 蚌科钩介幼虫的比较形态学研究Ⅱ.六个种幼虫的形态. 水生生物学报, 2000, 24(3): 252-256. DOI:10.3321/j.issn:1000-3207.2000.03.009] |
| [17] |
Wen HB, Ma XY, Xu P et al. Artificial selection of host fish and parasitic cyst formation of glochidia in pink heelsplitter, Potamilus alatus. Acta Hydrobiologica Sinica, 2018, 42(2): 356-363. [闻海波, 马学艳, 徐跑等. 紫黑翼蚌钩介幼虫寄主鱼的人工筛选及寄生包囊形成观察. 水生生物学报, 2018, 42(2): 356-363. DOI:10.7541/2018.045] |
| [18] |
Xu L, Wu XP, Ling G et al. Reproductive traits and glochidium morphology of Lamprotula leai(Gray). Journal of Nanchang University: Natural Science, 2013, 37(3): 262-266. [徐亮, 吴小平, 凌高等. 背瘤丽蚌繁殖特征及钩介幼虫形态. 南昌大学学报: 理科版, 2013, 37(3): 262-266. DOI:10.3969/j.issn.1006-0464.2013.03.012] |
| [19] |
Whelan NV, Geneva AJ, Graf DL. Molecular phylogenetic analysis of tropical freshwater mussels (Mollusca: Bivalvia resolves the position of Coelatura and supports a monophyletic Unionidae. Molecular Phylogenetics and Evolution, 2011, 61(2): 504-514. DOI:10.1016/j.ympev.2011.07.016 |
| [20] |
Pfeiffer Ⅲ JM, Graf DL. Evolution of bilaterally asymmetrical larvae in freshwater mussels (Bivalvia: Unionoida Zoological Journal of the Linnean Society, 2015, 175(2): 307-318. DOI: 10.1111/zoj.12282.
|
| [21] |
Lopes-Lima M, Froufe E, Do VT et al. Phylogeny of the most species-rich freshwater bivalve family (Bivalvia: Unionida modern subfamilies and tribes. Molecular Phylogenetics and Evolution, 2017, 106: 174-191. DOI:10.1016/j.ympev.2016.08.021 |
| [22] |
Bolotov IN, Vikhrev IV, Kondakov AV et al. New taxa of freshwater mussels (Unionidae) from a species-rich but overlooked evolutionary hotspot in Southeast Asia. Scientific Reports, 2017, 7: 11573. DOI:10.1038/s41598-017-11957-9 |
| [23] |
Huang XC, Su JH, Ouyang JX et al. Towards a global phylogeny of freshwater mussels (Bivalvia: Unionida) delimitation of Chinese taxa, mitochondrial phylogenomics, and diversification patterns. Molecular Phylogenetics and Evolution, 2019, 130: 45-59. DOI:10.1016/j.ympev.2018.09.019 |
| [24] |
Pfeiffer JM, Breinholt JW, Page LM. Unioverse: A phylogenomic resource for reconstructing the evolution of freshwater mussels (Bivalvia, Unionoida). Molecular Phylogenetics and Evolution, 2019, 137: 114-126. DOI:10.1016/j.ympev.2019.02.016 |
| [25] |
Lopes-Lima M, Bolotov IN, Do VT et al. Expansion and systematics redefinition of the most threatened freshwater mussel family, the Margaritiferidae. Molecular Phylogenetics and Evolution, 2018, 127: 98-118. DOI:10.1016/j.ympev.2018.04.041 |
| [26] |
Bolotov IN, Vikhrev IV, Bespalaya YV et al. Multi-locus fossil-calibrated phylogeny, biogeography and a subgeneric revision of the Margaritiferidae (Mollusca: Bivalvia Molecular Phylogenetics and Evolution, 2016, 103: 104-121. DOI: 10.1016/j.ympev.2016.07.020.
|
| [27] |
Araujo R, Schneider S, Roe KJ et al. The origin and phylogeny of Margaritiferidae (Bivalvia, Unionoida): A synthesis of molecular and fossil data. Zoologica Scripta, 2017, 46(3): 289-307. DOI:10.1111/zsc.12217 |
| [28] |
Huang XC, Rong J, Liu Y et al. The complete maternally and paternally inherited mitochondrial genomes of the endangered freshwater mussel Solenaia carinatus (Bivalvia: Unionidae) and implications for Unionidae taxonomy. PLoS One, 2013, 8(12): e84352. DOI:10.1371/journal.pone.0084352 |
| [29] |
Zhou CH, Ouyang S, Wu XP et al. Phylogeny of the genus Lamprotula (Unionidae) in China based on mitochondrial DNA sequences of 16S rRNA and ND1 genes. Acta Zoologica Sinica, 2007, 53(6): 1024-1030. [周春花, 欧阳珊, 吴小平等. 基于16S rRNA和ND1基因序列的中国蚌科丽蚌属的系统发育. 动物学报, 2007, 53(6): 1024-1030. DOI:10.3969/j.issn.1674-5507.2007.06.011] |
| [30] |
Song XL, Ouyang S, Zhou CH et al. Complete maternal mitochondrial genome of freshwater mussel Anodonta lucida (Bivalvia: Unionidae. Mitochondrial DNA Part A, 2016, 27(1): 549-550. DOI:10.3109/19401736.2014.905852 |
| [31] |
Wu RW, An CT, Wu XP et al. Complete maternal mitochondrial genome of freshwater mussel Aculamprotula tientsinensis (Bivalvia: Unionidae). Mitochondrial DNA Part A, DNA Mapping, Sequencing, and Analysis, 2016, 27(6): 4520-4521. DOI:10.3109/19401736.2015.1101543 |
| [32] |
Wang GL, Guo LP, Li JL. The F-type complete mitochondrial genome of Arconaia lanceolata. Mitochondrial DNA Part A, 2016, 27(1): 322-323. DOI:10.3109/19401736.2014.892098 |
| [33] |
Huang YY, Liu HZ, Wu XP et al. Testing the relationships of Chinese freshwater Unionidae (bivalvia)based on analysis of partial mitochondrial 16S rRNA sequences. Journal of Molluscan Studies, 2002, 68(4): 359-363. DOI:10.1093/mollus/68.4.359 |
| [34] |
Ouyang JX, Wu XP, Shan O et al. Phylogenetic analysis of some Chinese freshwater Unionidae based on mitochondrial COI sequences. Journal of Conchology, 2011, 40(5): 543-548. |
| [35] |
Ouyang JX, Su JH, Ouyang S et al. DNA barcoding and molecular phylogenetic analysis of freshwater unionids. Journal of Agricultural Biotechnology, 2015, 23(6): 779-787. [欧阳解秀, 苏金荟, 欧阳珊等. 蚌科DNA条形码识别与系统发育分析. 农业生物技术学报, 2015, 23(6): 779-787.] |
| [36] |
Wu RW, Liu XJ, Wang S et al. Analysis of mitochondrial genomes resolves the phylogenetic position of Chinese freshwater mussels (Bivalvia, Unionidae). Zoo Keys, 2019(812): 23-46. DOI:10.3897/zookeys.812.29908 |
| [37] |
Tamura K, Peterson D, Peterson N et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 2011, 28(10): 2731-2739. DOI:10.1093/molbev/msr121 |
| [38] |
Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution, 2013, 30(4): 772-780. DOI:10.1093/molbev/mst010 |
| [39] |
Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution, 2000, 17(4): 540-552. DOI:10.1093/oxfordjournals.molbev.a026334 |
| [40] |
Vaidya G, Lohman DJ, Meier R. Sequence Matrix: concatenation software for the fast assembly of multi-gene datasets with character set and Codon information. Cladistics, 2011, 27(2): 171-180. DOI:10.1111/j.1096-0031.2010.00329.x |
| [41] |
Zhang D, Gao FL, Jakovli c ' I et al. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources, 2020, 20(1): 348-355. DOI:10.1111/1755-0998.13096 |
| [42] |
Huang Y. Molecular Phylogenetics. Beijing: Science Press, 2012. [黄原. 分子系统发生学. 北京: 科学出版社, 2012.]
|
| [43] |
Lanfear R, Calcott B, Ho SYW et al. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution, 2012, 29(6): 1695-1701. DOI:10.1093/molbev/mss020 |
| [44] |
Kalyaanamoorthy S, Minh BQ, Wong TKF et al. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods, 2017, 14(6): 587-589. DOI:10.1038/nmeth.4285 |
| [45] |
Ronquist F, Teslenko M, van der Mark P et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 2012, 61(3): 539-542. DOI:10.1093/sysbio/sys029 |
| [46] |
Nguyen LT, Schmidt HA, von Haeseler A et al. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution, 2015, 32(1): 268-274. DOI:10.1093/molbev/msu300 |
| [47] |
Hu F, Lin Y, Tang J. MLGO: phylogeny reconstruction and ancestral inference from gene-order data. BMC Bioinformatics, 2014, 15: 354. DOI:10.1186/s12859-014-0354-6 |
| [48] |
Bernt M, Merkle D, Ramsch K et al. CREx: inferring genomic rearrangements based on common intervals. Bioinformatics, 2007, 23(21): 2957-2958. DOI:10.1093/bioinformatics/btm468 |
| [49] |
Drummond AJ, Suchard MA, Xie D et al. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution, 2012, 29(8): 1969-1973. DOI:10.1093/molbev/mss075 |
| [50] |
Bieler R, Mikkelsen PM, Collins TM et al. Investigating the Bivalve Tree of Life—an exemplar-based approach combining molecular and novel morphological characters. Invertebrate Systematics, 2014, 28(1): 32-115. DOI:10.1071/IS13010 |
| [51] |
Wu RW, Liu XJ, Ouyang S et al. Comparative analyses of the complete mitochondrial genomes of three Lamprotula (Bivalvia: Unionidae) species: insight into the shortcomings of mitochondrial DNA for recently diverged species delimitation. Malacologia, 2020, 63(1): 51-66. DOI:10.4002/040.063.0106 |
| [52] |
Graf DL, Jones H, Geneva AJ et al. Molecular phylogenetic analysis supports a Gondwanan origin of the Hyriidae (Mollusca: Bivalvia and the paraphyly of Australasian taxa. Molecular Phylogenetics and Evolution, 2015, 85: 1-9. DOI:10.1016/j.ympev.2015.01.012 |
| [53] |
Huang XC, Wu RW, An CT et al. Reclassification of Lamprotula rochechouartii as Margaritifera rochechouartii comb. nov. (Bivalvia: Margaritiferidae) revealed by time-calibrated multi-locus phylogenetic analyses and mitochondrial phylogenomics of Unionoida. Molecular Phylogenetics and Evolution, 2018, 120: 297-306. DOI:10.1016/j.ympev.2017.12.017 |
| [54] |
Wu RW, Liu YT, Wang S et al. Testing the utility of DNA barcodes and a preliminary phylogenetic framework for Chinese freshwater mussels (Bivalvia: Unionidae) from the middle and lower Yangtze River. PLoS One, 2018, 13(8): e0200956. DOI:10.1371/journal.pone.0200956 |
| [55] |
Wu RW, Wang S, Liu YT et al. Characterization and phylogenetic analysis of the complete maternal mitochondrial genome of freshwater mussel Aculamprotula scripta (Bivalvia: Unionidae). Conservation Genetics Resources, 2018, 10(4): 731-733. DOI:10.1007/s12686-017-0914-y |
| [56] |
Bolotov IN, Kondakov AV, Konopleva ES et al. A new genus of ultra-elongate freshwater mussels from Vietnam and Eastern China (Bivalvia: Unionidae). Ecologica Montenegrina, 2021, 39: 1-6. DOI:10.37828/em.2021.39.1 |
| [57] |
Cao YL, Liu XJ, Wu RW et al. Conservation of the endangered freshwater mussel Solenaia carinata (Bivalvia, Unionidae) in China. Nature Conservation, 2018, 26: 33-53. DOI:10.3897/natureconservation.26.25334 |
| [58] |
Dong ZG, Li XY, Li JL. Tolerance of alien species Hyriopsis schlegeli, native species Hyriopsis cumingii, and their hybrids to three ecological factors. Chinese Journal of Ecology, 2007, 26(7): 1080-1084. [董志国, 李晓英, 李家乐. 外来种池蝶蚌与本地种三角帆蚌及其杂交种对三种生态因子的耐受力. 生态学杂志, 2007, 26(7): 1080-1084. DOI:10.3321/j.issn:1000-4890.2007.07.021] |
| [59] |
Zhan XD, Wang SS, Huang YE et al. Species of freshwater mussels in Anhui Province. Journal of Jishou University: Natural Sciences Edition, 2018, 39(1): 65-67. [湛孝东, 王少圣, 黄月娥等. 安徽省淡水蚌类初步调查. 吉首大学学报: 自然科学版, 2018, 39(1): 65-67.] |
| [60] |
Xue TT, Liu XJ, Wu RW et al. Standing stock and spatial distribution pattern of unionids in Lake Taihu, China. J Lake Sci, 2019, 31(1): 202-210. [薛涛涛, 刘雄军, 武瑞文等. 太湖蚌类现存量及空间分布格局. 湖泊科学, 2019, 31(1): 202-210. DOI:10.18307/2019.0119] |
| [61] |
Wu RW, Gao BY, Lan ZC et al. Predicting local colonization and extinction rates of freshwater mussels based on biological traits in a case of Lake Poyang. J Lake Sci, 2017, 29(3): 678-686. [武瑞文, 高博宇, 兰志春等. 基于淡水蚌类的生物学特征预测种群局部定居率和灭绝率——以鄱阳湖为例. 湖泊科学, 2017, 29(3): 678-686. DOI:10.18307/2017.0317] |
| [62] |
Klishko OK, Lopes-Lima M, Froufe E et al. Unravelling the systematics of Nodularia (Bivalvia, Unionidae) species from eastern Russia. Systematics and Biodiversity, 2018, 16(3): 287-301. DOI:10.1080/14772000.2017.1383527 |
| [63] |
Ortmann AE. Notes upon the families and genera of the Najades. Annals of Carnegie Museum, 1912.
|
| [64] |
Modell H. Das natürliche System der Najaden. Archiv für Molluskenkunde, 1942, 74(5/6): 161-191. |
| [65] |
Heard WH, Guckert RH. A re-evaluation of the recent Unionacea (Pelecypoda) of North America. Malacologia, 1970, 10(2): 333-355. |
| [66] |
Zieritz A, Hoffman JI, Amos W et al. Phenotypic plasticity and genetic isolation-by-distance in the freshwater mussel Unio pictorum (Mollusca: Unionoida). Evolutionary Ecology, 2010, 24(4): 923-938. DOI:10.1007/s10682-009-9350-0 |
| [67] |
Hornbach DJ, Kurth VJ, Hove MC. Variation in freshwater mussel shell sculpture and shape along a river gradient. American Midland Naturalist, 2010, 164(1): 22-37. DOI:10.1674/0003-0031-164.1.22 |
| [68] |
Inoue K, Hayes DM, Harris JL et al. Phylogenetic and morphometric analyses reveal ecophenotypic plasticity in freshwater mussels Obovaria jacksoniana and Villosa arkansasensis (Bivalvia: Unionidae). Ecology and Evolution, 2013, 3(8): 2670-2683. DOI:10.1002/ece3.649 |
| [69] |
Zanatta DT, Murphy RW. Evolution of active host-attraction strategies in the freshwater mussel tribe Lampsilini (Bivalvia: Unionidae). Molecular Phylogenetics and Evolution, 2006, 41(1): 195-208. DOI:10.1016/j.ympev.2006.05.030 |
 2021, Vol. 33
2021, Vol. 33 

